


A ribosome is a cellular machine that makes proteins. Proteins have many different functions in a cell, ranging from forming structures to controlling chemical reactions. A human being uses tens of thousands of different kinds of proteins in his body, all made by ribosomes. According to evolutionary theory, a source of energy such as ultra-
Ribosomes are extremely complicated. A glance at the following animations shows just how this is so. The animations have great value in showing the impossibility for life to appear spontaneously from natural sources, as we shall see.
There is a fundamental, basic issue of contention between evolutionists and creationists. The very possibility of a natural origin of life depends on who is correct about this issue. On the one hand evolutionists propose that a structure such as a ribosome gradually acquired its current level of complexity through a series of very small changes, with each change providing increasingly effective performance. The idea is that each step of the series would become simple enough that it would be reasonably likely to occur within a reasonable period of time. Creationists on the other hand believe that a structure such as a ribosome needs to have a single-
There are four major problems that a ribosome presents to evolution:
1) A ribosome is composed of over 50 individual kinds of proteins, all precisely defined with precise functions. Yet, the only known way for a specific protein to form is from an already working ribosome. So, how can a ribosome gradually emerge on the basis of specific, step-
2) Scientists have never been able to form even a 50-
3) We will show that the proteins in a ribosome have very specific properties. This in turn requires that they have a very specific sequence of amino acids. How difficult would it be for a specifically needed protein of 100 randomly selected amino acids to provide a required sequence? It is unlikely for a single, isolated copy of it to appear in a single step on any hypothetical planet in the observable universe for just one hour in over 10,000,000,000,000,000,000,000,000,000 years. Even this would take conditions so favoring its appearance as to be extremely unrealistic and not applicable to real life. A trillion years is only 1,000,000,000,000 years. It is dwarfed by this number. This demonstrates how difficult it would be to get only a single isolated protein of 100 amino acids to appear anywhere in the universe for an just one hour. Is one hour too short? If you prefer to have available for 100 times as long (i. e., one hundred hours), then the time for it appear just increased by 100 times.
The problems against a natural appearance of a ribosome do not stop here. Eight of the proteins used in a ribosome are over 200 amino acids in length. We estimate that for every amino acid added to a chain, the odds against it being usable are about 1 in 10. The reasons for this are discussed in another article. This means that it would take about about an additional ten times as long for the required amino acid sequence to appear for each additional amino acid required. A 200-
In truth, if single-
4) Currently, scientists cannot even make a single RNA nucleotide in a laboratory using plausibly pre-
As one looks at the following animations, it would be well to keep in mind that the big issue concerns whether the observed data reasonably allows the possibility of gradual step-
Three Ribosome Animations That Debunk a Natural Origin of Life
By Timothy R. Stout


Ribosome Small Subunit
This is an animation of the small subunit of the ribosome used in e. coli bacteria. The animation demonstrates its 3-
As the subunit turns, notice the groove running across its width, about 1/3 down from the top. This groove provides the bottom portion of a channel through which a long molecule called messenger RNA travels as a new protein is fabricated. The information specifying the amino acid sequence of the protein being built is provided by messenger RNA. The messenger RNA conveys the information on how to build the protein stored in the DNA.
Notice the extreme complexity. A large number of proteins are used as structural elements to get the RNA to fold into its proper shape. Every single protein must have the exactly required shape and the exactly required selection of amino acids to bond properly to the RNA molecule at the correct location; otherwise, the entire assembly fails. The entire combination needs to make a single-
The small subunit independently self assembles within a cell. The various individual RNA molecules and proteins to be used are “dumped” into a cell unassembled. Then, when a certain, specific protein out of the group randomly comes into contact with the RNA molecule, it identifies a certain location on it and then attaches to that point. The electrical and chemical characteristics for this to happen need to be built into the construction of both the RNA molecule and the protein. Also, no other proteins in the cell and no other nucleotide sequences of the RNA can be allowed to have similar identification characteristics. Otherwise, false self assembly will occur. Once the protein has attached properly, the RNA/protein combination reconfigures into a new shape. The second ribosome protein is only able to attach to the newly acquired shape. Then, after the second protein has attached, the emerging ribosome takes on a yet new shape. This process repeats until all of the proteins have self assembled.
Self-
Evolutionists frequently talk as though emergence and self-
Therefore, self assembly adds yet another layer of complexity to the specification of a protein. Each protein not only needs to know where to assemble and what bond characteristics to exhibit to bond properly with the assembly at the appropriate time, it also needs to have the proper shape and bonding potential to preclude bonding improperly to any other potential structure that it comes in contact with. This includes not only the other components of the small subunit, but also of the large subunit, of the various synthetases, transfer RNAs, elongation factors, golgi body components, cellular spindle components, and a long list of other potential components. This, in truth, requires not only for the complete ribosome to be properly defined from the beginning, but also an entire “laundry list” of other components get included. In effect, the entire cell needs to make a single-
To keep this in the proper perspective, remember that it is unlikely to discover by chance a specifically required, 200-
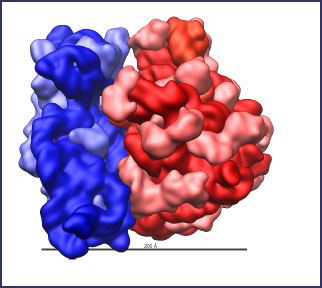
Ribosome Large Subunit
This is an animation of the large subunit of a ribosome of an e. coli bacterium. The orange, coiled strand is a single strand of RNA composed of a string of 2,906 nucleotides arranged in a specific order. The green is shadow. The yellow strand at the top is a second RNA strand composed of 120 nucleotides. The blue structures are proteins. A large subunit uses 4,223 amino acids spread among 36 different proteins. This is even more complex than the small subunit. However, it takes this degree of complexity for the ribosome to accomplish its task. The large subunit also self assembles.
The large subunit is much more complicated than the small one in that it has many more channels for various components to pass through. These channels must be precise in their individual shapes and locations and need to interconnect with each other in a precise manner. There is no room for experimenting with various possibilities until a suitable one appears. Either the ribosome works or it doesn’t. Natural selection cannot choose between the better of two failures. If a certain assembly fails to work, it is irrelevant to natural selection as to how close it might have been to working. There is no selection advantage to a failure.
Assembled Ribosome
This is an illustration of how a small subunit and large subunit appear when assembled together. When a small subunit has a messenger RNA properly located within its groove and the proper transfer RNA molecule attached to the messenger RNA, as shown in the next animation, the large subunit attaches to the lower subunit.
The attachment is a self-
Once fabrication of the new protein has been completed, the messenger RNA and the finished protein leave the ribosome , the lower subunit returns to its original shape, and the two subunits split apart. The capability of the small subunit to change shape in accordance with the status of the ribosome adds to the difficulties of getting a suitable assembly of nucleotides to fabricate it. The capability for the assembled ribosome to change its shape as necessary is a critical function which needs to be present from its beginning.
So, the various proteins of the ribosome not only need to be configured in such a way that they can self-

How a Ribosome Works
In the animation to the right, the lower yellow structure is a ribosome small subunit. The black thread extending across the page is a strand of messenger RNA. It will function as a template containing the information to specify the choice of amino acids in fabricating through protein to be built. When a messenger RNA molecule and a small subunit of a ribosome cross paths, the messenger RNA will attach to the small subunit in its groove. There is a mechanism to get everything aligned properly.
The components with the bright blue structures are called transfer RNA. A transfer RNA molecule has an amino acid attached to it at its tip. At its base it has identifier “codons” which determine whether it represents the kind of amino acid needed at this spot in the emerging protein. The amino acid attached to the tip will ultimately be transferred from the transfer RNA to the proper location in the emerging protein. A protein is composed of an assortment of 20 different amino acids. Therefore, a cell has at least 20 different kinds of transfer RNAs, one for each kind of amino acid.
Only the kind of tRNA specified by the messenger RNA will bond to it at any given bonding site. The light blue attachment to the tRNA is called a synthetase. All the different kinds of transfer RNA have the same basic structure where the amino acid is attached. A separate molecule called a synthetase actually joins the proper amino acid to the proper transfer RNA. There is a channel between the small subunit and the large subunit for a transfer RNA to enter a ribosome. If the code information at the bottom of the transfer RNA matches the information in the messenger RNA telling what kind of amino acid is needed, the transfer RNA bonds to the messenger RNA and the synthetase is released. Otherwise (not shown in the animation) the transfer RNA is rejected, leaves the ribosome, and a new one enters. Rejection is the normal outcome. With twenty choices of amino acids and only one correct one, most of the time the wrong amino acid will appear.
A ribosome can hold three transfer RNAs at a time. The center one attaches its amino acid to the emerging string of amino acids forming the new protein. The one entering the ribosome with its synthetase attached will supply the next amino acid of the protein.
Once the synthetase has been released from the ribosome, indicating that the proper transfer RNA has bonded to the messenger RNA, a large protein called an “elongation factor” attaches to the ribosome and pushes the entire messenger RNA/transfer RNA assembly forward. In a human being, the elongation factor is composed of 578 amino acids. How many multiple Googles of years should it take to get a single, momentary appearance of this protein from random processes?
The animation above shows the transfer RNA changing shape as it advances from the first to the middle position. This shape change pushes the emerging protein through a small channel until it exits out the top of the ribosome. This shape change is a tricky process. The transfer RNA molecule needs to rise the proper amount to advance the protein the exactly required amount so that the next amino acid to be attached will line up properly. The amino acid sequence of the transfer RNA needs to change its shape in accordance with its progress through the ribosome. This is another characteristic which needs to appear correctly at its first time; otherwise, the amino acids do not get properly joined to each other.
The remaining steps shown in the animation concern activities related to protein processing subsequent to the actual protein assembly and will not be discussed here. However, it is obvious that the structures are much more complicated than an elongation factor and that is composed of well over five hundred amino acids.
There is another difficulty. ATP (adenosine tri-
It is difficult to believe that sunlight or some equivalent unconstrained source of energy could convert raw materials such as methane, ammmonia, carbon dioxide, hydrogen, water, etc. into a system with this degree of complexity. Also, the things we have looked at give strong evidence that the entire first cell needed to make a single-
Ribosome Small Subunit
Ribosome Large Subunit
Assembled Ribosome
How a Ribosome Works
Concluding Remarks
If one works through everything required to build properly a ribosome, it seems clear that it needs to make a single-
Natural processes naturally go one way. By contrast, a living cell needs specific chemicals at specific times and locations. There is nothing to constrain or restrain what is produced to what is needed. As a result, the wrong things are produced and a natural origin of life cannot appear. This is the hypothesis of Abiogenetic Disconnects talked about elsewhere on this website.
Evolutionists talk as if the use of RNA in a ribosome is evidence of an earlier instance of what is called an “RNA World.” It makes more sense to view the combination of RNA and protein as the most efficient way to effect the self-
Ultra-
Since the preceding discussion gives strong, observable evidence favoring a single-
If natural processes are all that are available, according to what we have learned from science we shouldn’t exist. However, we do. Hence, we need to have an origin outside of natural processes. Thus, an honest appraisal of the observations of science provide strong evidence favoring special creation and not chemical evolution as the source of life.
The above animations were taken from the wikipedia.org article on “Ribosome.”
The animations are used in accordance with the
It is recommended that our article Random Sequence Difficulties be read together with this one, as the two supplement each other in providing a single argument about the difficulty and significance of life emerging from random sequences. The “Difficulties” article goes through calculations showing just how unlikely it would be for natural processes to make the chemicals of life. The results are staggering. For most chemicals, the entire universe is very unlikely to form just one instance of only one of them in a Googol years. It takes an already living cell to make these chemicals. This article shows single-
Counter installed November 14, 2016.
Only the first access per reader counts.